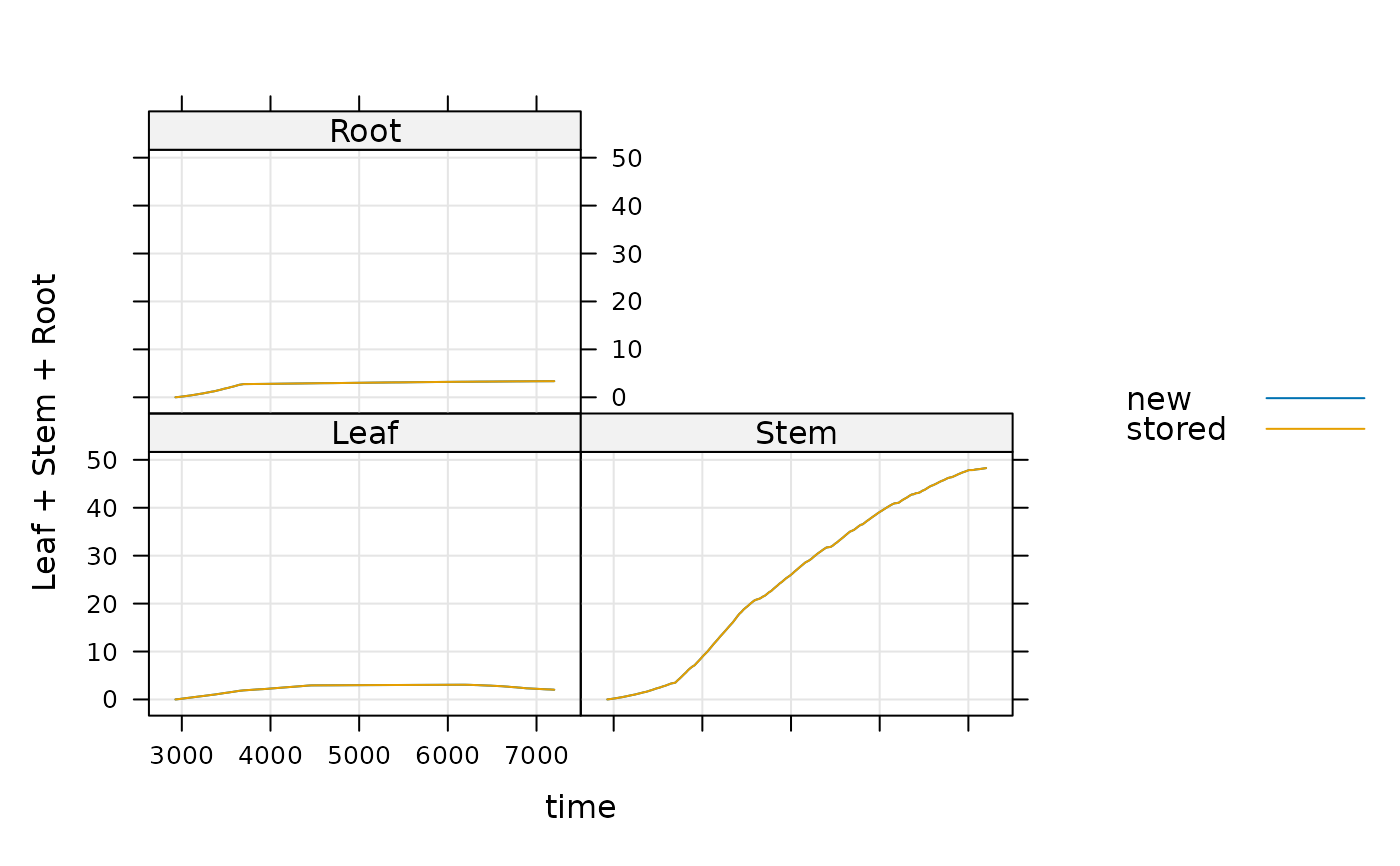
Compare new and stored results for a BioCro model test case
compare_model_output.RdBioCro models can be tested using test cases, which are sets of known outputs
that correspond to particular inputs. The compare_model_output
function facilitates manual comparisons between new and stored results.
Note that model tests are distinct from the module tests
described in module_testing.
Arguments
- mtc
A single module test case, which should be created using
model_test_case.- columns_to_keep
A vector of column names that should be included in the return value. If
columns_to_keepisNULL, all columns that are in both the new and stored result will be included.
Details
The compare_model_output function is a key part of the BioCro
model testing system. See model_testing for more information.
This function will run the model to get a new result, and load the stored
result associated with the test case. The two data frames will be combined
using rbind, where a new column named version indicates
whether each row is from the new or stored result.
It is intended that quantities from the resulting data frame will be plotted to visually look for changes in the model output.
Examples
# Define a test case for the miscanthus model and save the model output to a
# temporary directory
miscanthus_test_case <- model_test_case(
'miscanthus_x_giganteus',
miscanthus_x_giganteus,
get_growing_season_climate(weather$'2005'),
TRUE,
tempdir()
)
update_stored_model_results(miscanthus_test_case)
# Now we can use `compare_model_output` to compare the saved result to a new one
comparison_df <- compare_model_output(miscanthus_test_case)
# This will be a boring example because the new and stored results will be
# exactly the same
lattice::xyplot(
Leaf + Stem + Root ~ time,
group = version,
data = comparison_df,
type = 'l',
auto = TRUE,
grid = TRUE
)